NeuroDataPub Assistant Guide
Important
Before using NeuroDataPub, the remote data server should provide at least an installation of git-annex. Please see Remote Data Server Setup for instructions.
Note also that NeuroDataPub takes as principal input the path of your dataset that should be compliant to the Brain Imaging Data Structure (BIDS) format
by default.
If you are using a dataset in BIDS format, you should always make sure that your dataset is in valid BIDS format before using NeuroDataPub using
the free, online BIDS Validator, or its standalone version.
See BIDS standard for more information about BIDS.
If it does not make any sense to adopt the BIDS format for your dataset, NeuroDataPub can also handle dataset not necessary in the BIDS format,
since v0.4, with the --is_not_bids option.
Introduction
NeuroDataPub comes with a Graphical User Interface
aka the NeuroDataPub Assistant to support not only
the configuration of the siblings and the generation of the
corresponding JSON configuration files, but also its
execution in the three different modes.
1. Start the Graphical User Interface
In a terminal, activate the neurodatapub-env conda environment:
$ conda activate neurodatapub-env
Please check Creation of neurodatapub-env conda environment for more details about its creation.
After activation, the NeuroDataPub Assistant can be launched
via the neurodatapub command-line interface with the --gui option flag:
$ neurodatapub --gui \ (--dataset_dir '/local/path/to/input/bids/dataset' \) (--datalad_dir '/local/path/to/output/datalad/dataset' \) (--git_annex_ssh_special_sibling_config '/local/path/to/special_annex_sibling_config.json' \) (--github_sibling_config '/local/path/to/github_sibling_config.json') (--osf_sibling_config '/local/path/to/osf_sibling_config.json')
Note
When you run the neurodatapub command-line interface with the --gui option, it is not required to
specify the option flags required for a normal run from the commandline interface.
However, if provided, the parameters will be used to initialize the configuration of the project.
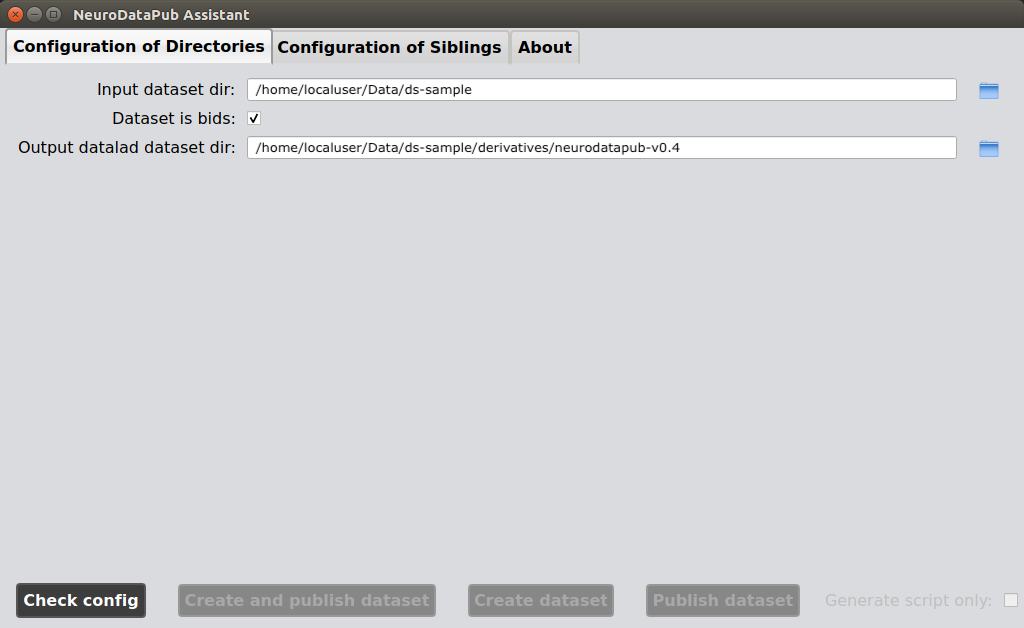
2. Configure input and outputs directories
You can select or reconfigure your input dataset directory, its format (BIDS / non-BIDS) and the directory of the
Datalad dataset that will be created in the first tab of the NeuroDataPub Assistant.
3. Configure the siblings
You can configure or reconfigure the settings for the special git-annex and GitHub remote siblings.
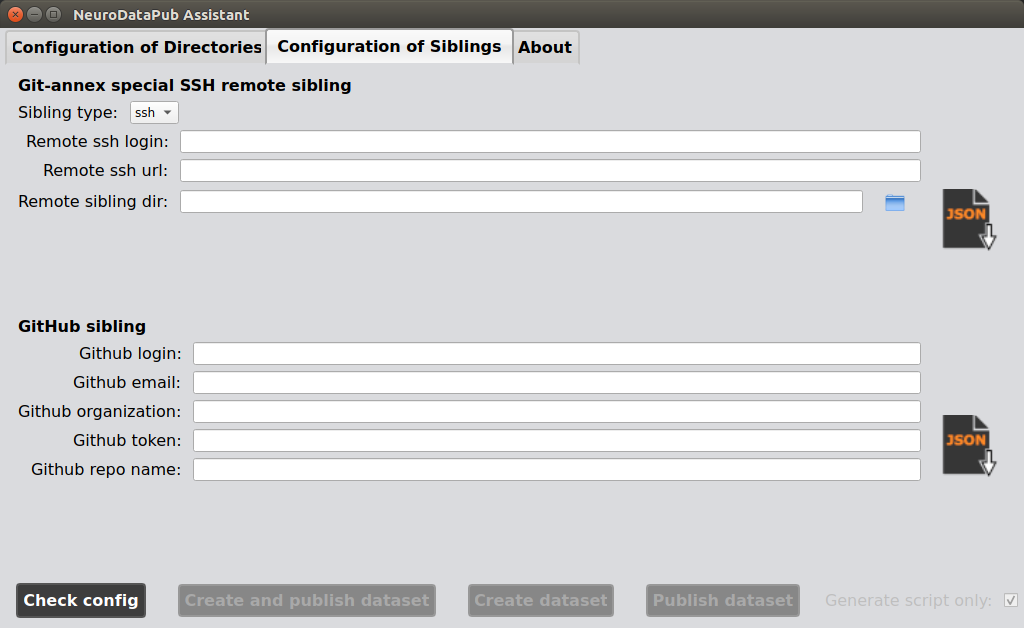
3.1 Special remote sibling settings
Since v0.3, you can use either (1) the data storage server of your institution accessible
via ssh or (2) the Open Science Foundation (OSF) platform to host your annexed files.
3.1.1 Server accessible via ssh
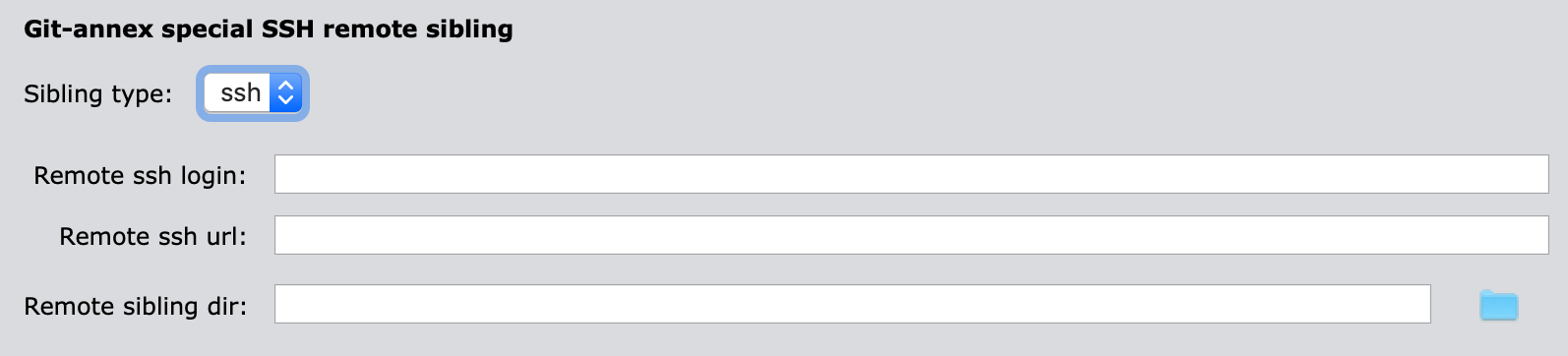
"remote_ssh_login"(mandatory): user’s login to the remote"remote_ssh_url"(mandatory): SSH-URL of the remote in the form"ssh://...""remote_sibling_dir"(mandatory): Remote .git/ directory of the sibling dataset
3.1.2 OSF (Cloud)
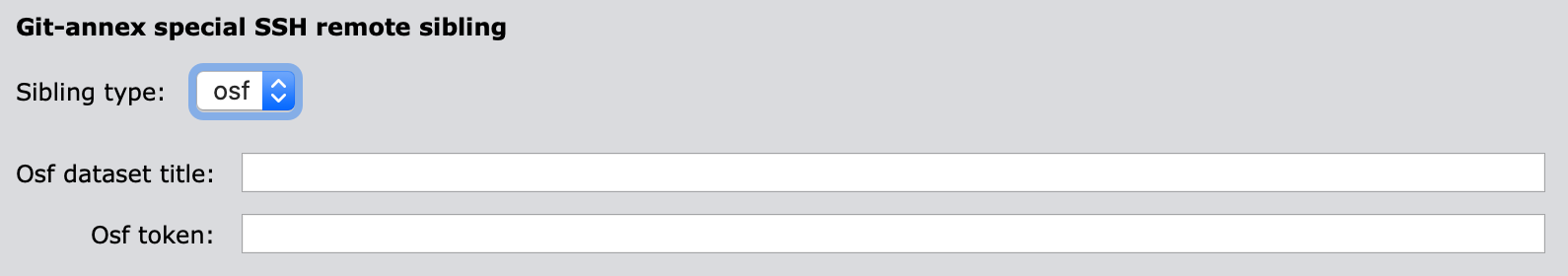
"osf_dataset_title"(mandatory): Dataset title on OSF."osf_token"(mandatory): user’s OSF authentication token. To make a Personal Access Token, please go to the relevant OSF settings page and create one. If you do not an OSF account yet, you will need to create one a-priori.
3.2 GitHub sibling settings
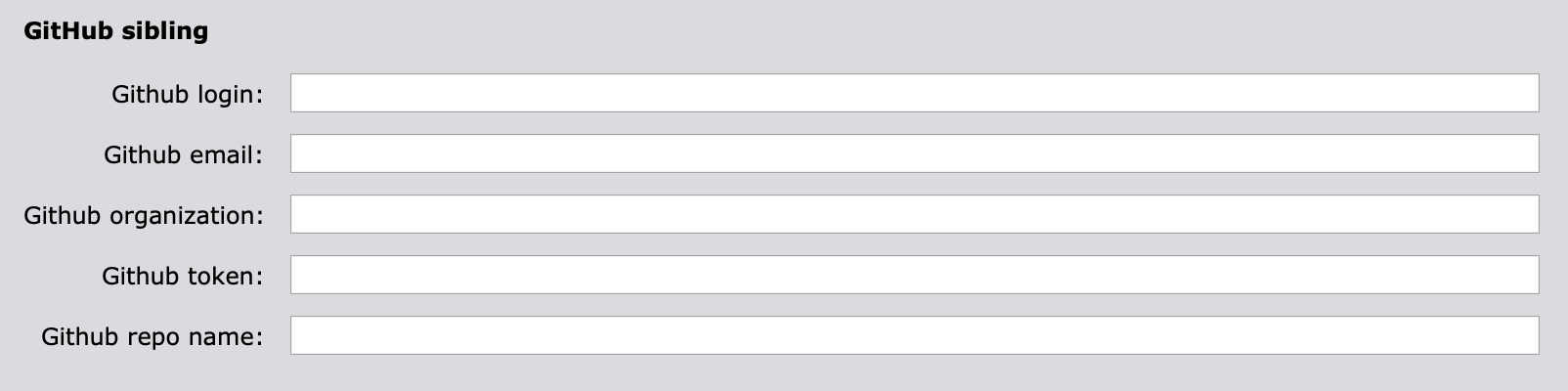
"github_login"(mandatory): user’s login to GitHub."github_email"(mandatory): user’s email associated with GitHub account."github_organization"(mandatory): GitHub organization the GitHub account has access to."github_token"(mandatory): user’s github authentication token. Please see “Creating a personal access token” Github documentation for more details on how to get one. Make also sure that thewrite:organdread:orgoptions are enabled."github_repo_name"(mandatory): Dataset repository name on GitHub.
3.3 Create the JSON sibling configuration files
Settings for each of the different siblings can be saved in a JSON file by clicking
on their respective ![]() button.
button.
4. Check the configuration and run NeuroDataPub
Before being able to initiate the processes of creation and / or publication
of the datalad dataset, you will need to make the NeuroDataPub Assistant
checking them out by clicking on the Check config button.

If the configuration is completely valid, this will enable the
Create and Publish Dataset, Create Dataset, Publish Dataset buttons.

Then, you can run NeuroDataPub in one of the three execution modes by clicking on one of the
buttons.

Need more control?
Since v0.4, NeuroDataPub can be run in Generate script only mode to give more control to more advanced users familiar with the Linux shell.
If enabled, NeuroDataPub will run in a “dryrun” mode and will only create a Linux shell script called neurodatapub_%d-%m-%Y_%H-%M-%S.sh in the code/
directory of your input dataset that records all the underlined commands. If it appears that the code/ folder does not exist yet, it will be
automatically created.

Note
You can always see the execution progress by checking the standard outputs in the terminal, such as the following:
$ neurodatapub --gui
[...]
############################################
# Check configuration
############################################
* PyBIDS summary:
BIDS Layout: ...localuser/Data/ds-sample | Subjects: 1 | Sessions: 1 | Runs: 0
* remote_ssh_login: user
* remote_ssh_url: ssh://stockage.server.ch
* remote_sibling_dir: /home/user/Data/ds-sample/.git
* github_login: user
* github_repo_name: ds-sample
Configuration is valid!
############################################
############################################
# Creation of Datalad Dataset
############################################
> Initialize the Datalad dataset /home/localuser/Data/ds-sample/derivative/neurodatapub-v0.1
[INFO ] Creating a new annex repo at /home/localuser/Data/ds-sample/derivative/neurodatapub-v0.1
[INFO ] Running procedure cfg_text2git
[INFO ] == Command start (output follows) =====
[INFO ] == Command exit (modification check follows) =====
[INFO ] Running procedure cfg_bids
[INFO ] == Command start (output follows) =====
[INFO ] Running procedure cfg_metadatatypes
[INFO ] == Command start (output follows) =====
[INFO ] == Command exit (modification check follows) =====
[INFO ] == Command exit (modification check follows) =====
Dataset(/home/localuser/Data/ds-sample/derivative/neurodatapub-v0.1)
[...]
Support, bugs and new feature requests
All bugs, concerns and enhancement requests for this software are managed on GitHub and can be submitted at https://github.com/NCCR-SYNAPSY/neurodatapub/issues.